On December 11, the Laboratory of Structural Biochemistry at Shanghai Institute for Advanced Immunochemical Studies (SIAIS), led by Distinguished Adjunct Professor Roger Kornberg and Research Associate Professor Zhang Heqiao, published a research article in Science Advances titled “Cryo-EM Structure of Nipah Virus RNA Polymerase Complex.” This study provides a detailed characterization of the structural features of the Nipah virus polymerase complex.
The Nipah virus is a member of the non-segmented, negative-sense RNA viruses (nsNSV) family, which includes several other highly pathogenic viruses such as Ebola virus, Marburg virus, and mumps virus. Nipah virus, first identified in Malaysia in 1998, belongs to the Paramyxoviridae family. Over time, it has rapidly mutated and spread to other Asian regions, including Singapore, Bangladesh, India, Cambodia, and China’s Langya region in Shandong Province. In recent years, Nipah virus infection cases have been intermittently reported in Asia. The virus is primarily transmitted through animal-to-human and human-to-human contact, and the infection causes severe symptoms such as encephalitis and pneumonia, with a fatality rate of over 70%. Currently, there are no approved vaccines or antiviral drugs for its prevention or treatment, as development efforts are still ongoing. The Nipah virus genome encodes six proteins, among which the RNA-dependent RNA polymerase complex (composed of the L and P proteins) is an attractive target for antiviral drug development.
In the recent study, the team successfully overexpressed the Nipah virus polymerase complex in insect cells, purified it to homogeneity, and determined its structure at 2.9 Å resolution using single-particle cryo-electron microscopy (Cryo-EM) (Figures 1A and 1B). Similar to other nsNSVs, the Nipah virus P protein forms a tetramer and assembles with the L protein through multiple interfaces to form an entire polymerase complex. The team also identified the entrance and exit tunnels for template RNA, product RNA, and nucleotide triphosphates (NTPs) on the polymerase structure. Intriguingly, structural superposition with template RNA-bound Ebola virus and respiratory syncytial virus revealed that the nsNSV family shares several conserved key amino acids involved in template-RNA recognition (Figure 1C). This finding suggests that the nsNSV family likely employs a highly conserved mechanism for template-RNA recognition. In addition, the mutation-prone sites among Nipah virus strains have been mapped onto the structure (Figure 1D), providing an important structural basis for the future development of antiviral drugs and vaccines.
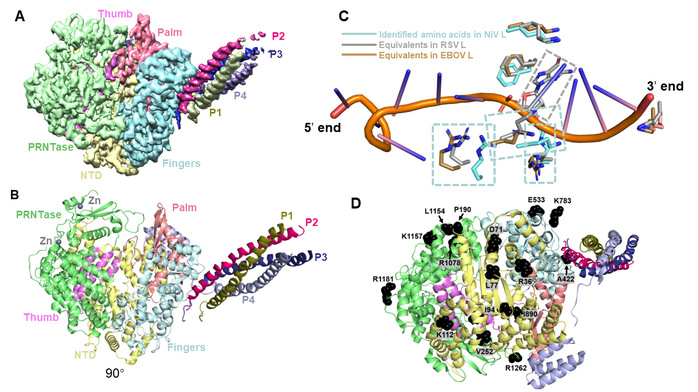
Figure 1. Structural features of the NiV polymerase complex. (A) Cryo-EM reconstruction map of the NiV polymerase complex. (B) Cryo-EM structure of the NiV polymerase complex. (C) Conserved recognition mode for template RNA employed by nsNSVs. (D) Mapping of the mutation-prone sites onto the polymerase structure.
Co-first authors of the study include third-year PhD candidate Wang Yiru, senior engineer Zhao Lixia, engineer Zhang Yi from SIAIS, and third-year master’s student Wang Yuhan from the iHuman Institute. The co-corresponding authors are Zhang Heqiao and Roger Kornberg from SIAIS, along with Luca Zinzula and Zhang Xiaoxiao from the iHuman Institute. ShanghaiTech University is the primary affiliation, and all cryo-EM data were collected at the Bio-Electron Microscopy Facility.
*This article is provided by Zhang Heqiao

