A research team led by Assistant Professor Yang Bei at the School of Life Science and Technology (SLST) and Shanghai Institute for Advanced Immunochemical Studies (SIAIS) and Professor Chen Jia at SLST, ShanghaiTech University, has achieved a significantprogress in mitochondrial DNA (mtDNA) editing. Their study, published in Molecular Cell, revealed the first structural snapshots of functioning DdCBE, a mitochondrial base editor. The researchers developed WinPred model and adDCBE to enable single-nucleotide editing of mtDNA and precise modeling of disease-related mtDNA mutations.
Mutations in mtDNA cause a range of severe multisystem disorders, particularly in high-energy-demanding tissues like the central nervous system. For example, Leber hereditary optic neuropathy (LHON), can be caused by homoplasmic (all mtDNA copies carry the mutation), point mutations in mitochondrial genes like NADH dehydrogenase 1 gene (MT-ND1), which mainly affect the optic nerves and lead to central vision loss. However, existing CRISPR-derived gene-editing technologies cannot be used to correct such mutations as guide RNAs cannot enter mitochondria, while programable nucleases risk destroying all copies of the mitochondrial genome, leading to catastrophic consequences. To enable correction of mtDNA mutations, researchers developed mtDNA-targeting base editors. The first, DdCBE, couples transcription-activator-like effector (TALE) arrays with the dsDNA-specific cytidine deaminase DddA to target specific mtDNA loci for C-to-T editing. Nevertheless, DdCBE and its derived variants exhibit elusive editing windows and non-negligible off-target (OT) effects, limiting their application in high-precision-demanding research and clinical settings.
To address these challenges, the team first determined high-resolution cryo-EM structures of DdCBE targeting two endogenous mtDNA loci, thus revealing its working mechanism and the structural determinants of its editing window (Graphical summary, top panel, left). Integrating biochemical and cellular editing data, they then established WinPred, a model capable of predicting editing windows across different spacer lengths (Graphical summary, top panel, right). Guided by WinPred, the design of TALE-recognition regions and spacer lengths of sDdCBEs can be optimized to achieve strand-specific, high-precision editing at different mtDNA loci. Building further on structural insights, they engineered the DddA effector domain to create an accurate version of DdCBE (aDdCBE) with a narrowed 2–3 nt editing window and near-background level off-target effects (Graphical summary, bottom panel). By combining WinPred and aDdCBE, single-nucleotide precision mtDNA editing can be achieved even at challenging target site, where target cytosine is flanked by multiple bystander cytosines (Graphical summary, middle panel, left). Using this system, the team precisely introduced a pathogenic mutation associated with LHON into mtDNA, faithfully reproducing the disease phenotype and demonstrating the potential of WinPred and aDdCBE for accurate modeling of mitochondrial disorders (Graphical summary, middle panel, right).
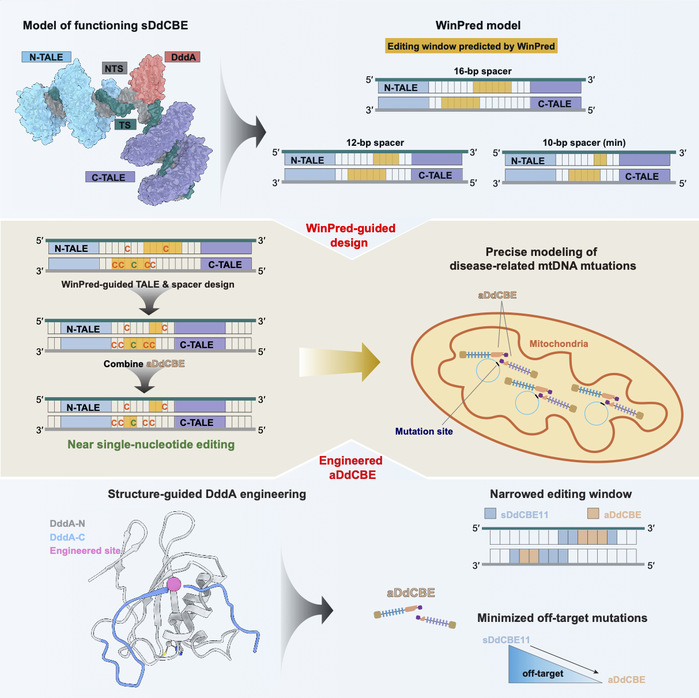
Graphical summary
This study provided the first structural view of DdCBE in action, offering critical mechanistic insights into its editing window and laying a theoretical foundation for future optimization of mitochondrial base editors. The newly developed WinPred model and aDdCBE system further provided powerful new tools for the precise modeling and potential therapeutic targeting of mitochondrial diseases.
Xiang Jiangchao from SIAIS and Xu Wenchao and Wu Jing from SLST are the co-first authors. Yang Bei and Chen Jia are the co-corresponding authors. ShanghaiTech University is the first affiliation.
*This article was is provided by Prof. Yang Bei

